
Applied Science Letters


Increased globalization and trade lead to the evolution of several viral pathogens which acclimatize to new hosts and augment their habitat alteration are some factors responsible for their emergence. Hantaviruses are upcoming viruses that are transmitted by small Rodents especially Rats. Once communicated to humans, they can result in two important clinical syndromes, hemorrhagic fever with renal syndrome, and Hantavirus cardiopulmonary syndrome. Humans are inadvertent hosts generally got infected through aerosols produced from infected rodents contamination by urine, feces, and saliva. The infections spread from humans to humans are rare as the evolution of the virus imperatively relies on the rodent host. Hantaviruses are the members of the family Bunyaviridae/hantaviridae and commonly regarded as orthohantavirus (or hantavirus) which is a single-stranded, enveloped, negative-sense RNA virus belonging to the order Bunyavirales. Research on Hantavirus is carried out globally but at a slower rate. In the present scenario, the growth of the coronavirus has instigated to study on the Hantavirus and eradicate the growth of viruses with the latest vaccines. Our current review focuses on the pathophysiology and molecular mechanism of Hantavirus growth and measures recommended for the safety of human society.
Globally Emerging Hantavirus Clinical Syndromes and Mechanism to Eradicate the Threat to Human Society-A Short Review
K. R. Padma1*, K. R. Don2, P. Josthna1, V. Bindu3
1 Assistant Professor, Department of Biotechnology, Sri Padmavati Mahila VisvaVidyalayam (Women’s) University, Tirupati, AP, India.
2 Reader, Department of Oral Pathology, Saveetha Dental College, Saveetha Institute of Medical and Technical Sciences, Saveetha University, Velappanchavadi, Chennai, Tamil Nadu, India.
3 Assistant Professor, Department of HomeScience, Sri Padmavati Mahila VisvaVidyalayam (Women’s) University, Tirupati, AP, India.
ABSTRACT
Increased globalization and trade lead to the evolution of several viral pathogens which acclimatize to new hosts and augment their habitat alteration are some factors responsible for their emergence. Hantaviruses are upcoming viruses that are transmitted by small Rodents especially Rats. Once communicated to humans, they can result in two important clinical syndromes, hemorrhagic fever with renal syndrome, and Hantavirus cardiopulmonary syndrome. Humans are inadvertent hosts generally got infected through aerosols produced from infected rodents contamination by urine, feces, and saliva. The infections spread from humans to humans are rare as the evolution of the virus imperatively relies on the rodent host. Hantaviruses are the members of the family Bunyaviridae/hantaviridae and commonly regarded as orthohantavirus (or hantavirus) which is a single-stranded, enveloped, negative-sense RNA virus belonging to the order Bunyavirales. Research on Hantavirus is carried out globally but at a slower rate. In the present scenario, the growth of the coronavirus has instigated to study on the Hantavirus and eradicate the growth of viruses with the latest vaccines. Our current review focuses on the pathophysiology and molecular mechanism of Hantavirus growth and measures recommended for the safety of human society.
Keywords: Orthohantavirus, Negative sense RNA virus, Coronavirus, Human society, Hantavirus, Bunyavirales.
INTRODUCTION
Viral infections are considered as one of the principal threats to human life and health worldwide [1-3]. Worldwide outbursting of zoonotic pathogens continuous as a vital public threat arising from a various health problem. In the present scenario of Coronavirus threat to human society and social distancing, wakefulness for all viruses appreciated. In this regard, Hantaviruses was earlier causing chaos and at present also has developed a lot of attention as novel pathogenic in several parts of Asia, America, and Europe. Hantaviruses commonly infect small mammals and humans infected through contaminated aerosols or might be through contact with the droppings of small rodents. [4] Once being transmitted from the small rodents, the virus is responsible for major clinical syndromes such as Haemorrhagic fever with renal syndrome (HFRS), especially occurring in Asia as well as Europe and also causes hantavirus cardiopulmonary syndrome (HCPS/HPS) among the Americas. [5] Generally, after the investigations it has been recognized as Hantavirus and its first major outbreak was isolated by Ho Wang Lee near the Hantan River area in South Korea in 1976.[6]
The Hantavirus has three important genome segments such as S, M, and L RNA, respectively. The virion possessing the genome segments is encapsulated with nucleocapsid named (N) protein which is a glycoprotein precursor (GPC) to form ribonucleoprotein (RNP), along with RNA dependent RNA-polymerase (RdRp) protected within a lipid envelope with titivated spikes made of glycoproteins Gn and Gc. [7] The S RNA encodes the nucleocapsid (N) protein. The M RNA is cleaved to yield the envelope glycoproteins G1 and G2. Simultaneously the L RNA functions as the viral transcriptase/replicase enzyme. Therefore, the N protein provides a sufficient pool for mRNA transcription and translation and helps to initiate the process of replication. [8] Nevertheless, trimers of the N protein are critically obligatory for effectual translation in eukaryotic cells. [9]
Moreover, the glycoprotein expressions of the pathogenic hantaviruses hinder INF-β and TBK-1 induction by their virulence determinants existing on the G1 cytoplasmic tail. [10] Moreover, the N proteins can also affect the dimerization of protein kinase R (PKR) as well as the N protein inhibits PKR phosphorylation, which is crucial for its enzymatic activity. The PKR action is to inhibit virus replication and maintain antiviral state. The PKR's role is to trigger IFN via NF-κB and IRF1 up-regulation. [11] Therefore, in our current review, we will have acknowledged the N protein’s role in the regulation of antiviral state and emphasized the transmittance life cycle, briefly portrayed about vaccines currently available as well as depicted about the use of conventional therapy for prevention of viral disease.
Hantavirus Genome organization and its structure
The orthohantaviruses are enveloped viruses with a size of 80-120nm. Furthermore, the genome organization contains three negative-sense single-stranded RNA segments designated as S, M, and L protein which are encoded by the nucleocapsid (N) protein. The M RNA is multiprotein which in turn is directed for the process of translational and later hewed to produce the glycoproteins G1 and G2 which is embedded in the lipid envelope. The L RNA contains the enzymes' viral transcriptase/replicase for replication purposes. The Hantavirus belongs to the order Bunyavirales and each of the three segments possesses consensus 3'-terminal nucleotide sequence (AUCAUCAUC) which is complementary to the 5'-terminal sequence and is dissimilar from other four genera within the family. [12] Although each sequences segments i.e L protein is approximately 6530–6550 nucleotides bps in length, the M protein is 3613–3707 bps in length and the S protein is nearly 1696–2083 bps in length.
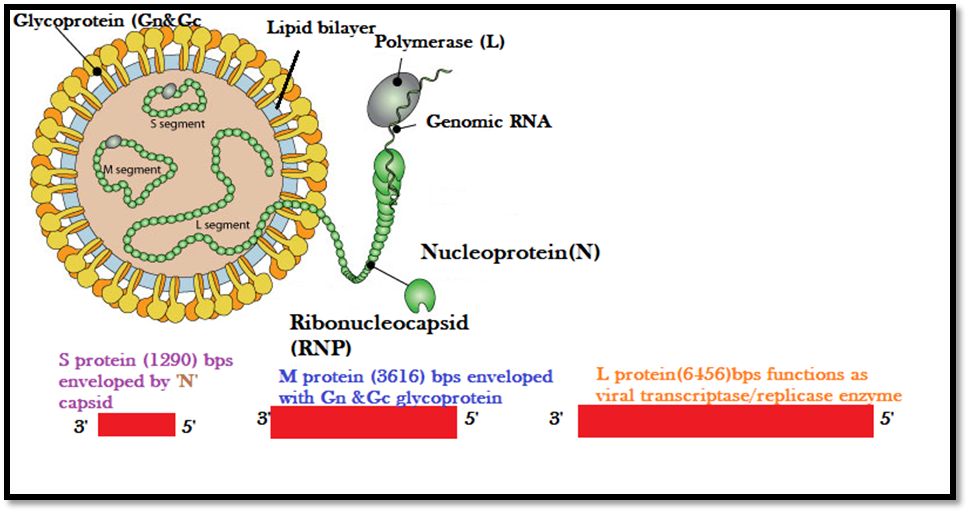
Figure 1: Hantavirus Genome Organization with an outer decorated Lipid envelope and genome protected within with Nucleocapsid regarded as N proteins.
Life cycle of Hantavirus transmittance
The important members of the genus Bunyaviridae include arboviruses and Hantaviruses which are maintained in particular rodent hosts (roboviruses). The distribution of Hantavirus specifically occurs in those regions where the reservoir rodent host exists. [13] Furthermore the natural reservoir of Hantaviruses is murid rodents belonging to the order Rodentia and family Muridae which has been cleaved into subfamilies Murinae, Arvicolinae, and Sigmodontinae. Yet it has not been revealed that viruses exist in non-rodent hosts for transmission of Hantaviruses.
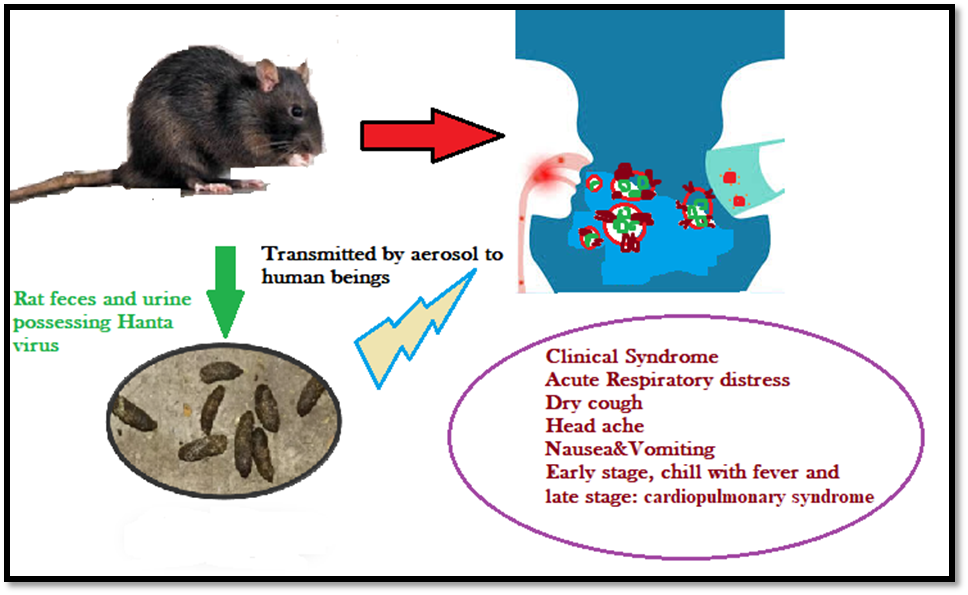
Figure 2: Schematic diagram depicting the Life cycle transmittance of Hantavirus
Glycoprotein Trafficking
To enter the host cell, the virus has to cross the cellular membrane barrier. During the process of entry, the Hantavirus glycoproteins play a key role by interacting with the cell surface receptors and trickily escape from the endocytic pathway through the fusion of viral and cellular membranes. [14] A schematic diagram that summarized the glycoprotein interaction and trafficking process as shown in (Figure-3). These viruses can bind to certain specific receptors on the cellular surface, such as β3 integrins, decay-accelerating factor (DAF)/CD55 as well as the receptor for the globular head domain of complement C1q (gC1qR)/p32. [15] Once the virus binds on the cellular membrane, the multiple receptors clustered on the membrane and thereby enhancing the acidity as well as activating the signaling process by introducing invagination to form an endocytic vesicle. [16]
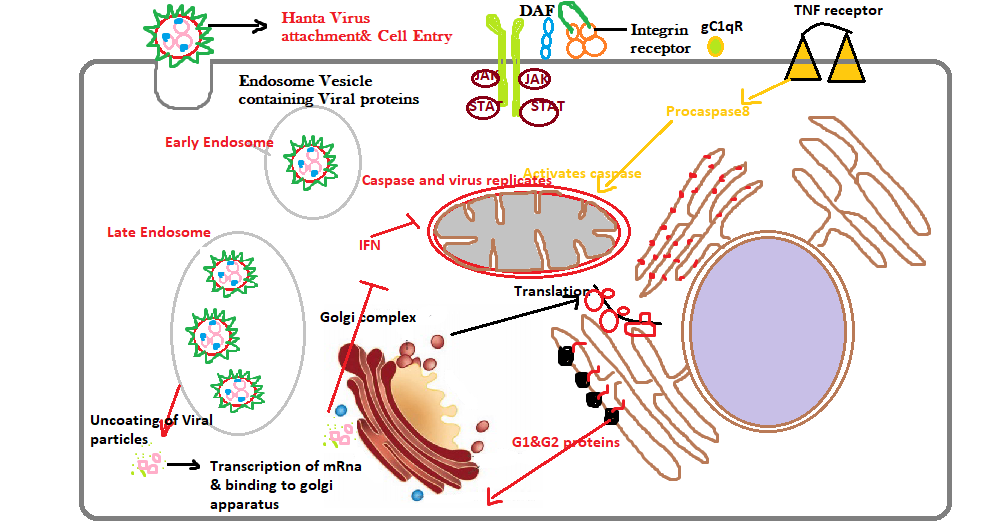
Figure 3: Hantavirus regulation mechanism with RNA binding and Ribonucleoproteinassembly during the trafficking process in the Golgi complex.
Additionally, the Hantavirus can infect not only the macrophage cells but as well as epithelial, endothelial, follicular dendritic, as well as lymphocyte cells via the viral glycoprotein attachment to the host’s cell surface receptors. [17] Various literature studies reported that the β-receptors especially interact with the viral glycoproteins G1 and G2, responsible for causing pathogenic syndromes such as HFRS and HPS. The basic mechanism of action in trafficking displayed the formation of endolysosomal cells, where the virus starts uncoating to release the three important RNPs into the cytoplasm. Once the Viral RdRp released, initiation of primary transcription takes place and later translation of the S, as well as L mRNA transcripts, occurs basically on free ribosomes. Amongst, the third protein named the M-segment transcript which is present on membrane-bound ribosomes, and gets matured on the rough endoplasmic reticulum. Nevertheless, in the case of Hantaviruses, the N protein is the most copious viral protein which is produced in abundance during the early stage of infection. Thus, all three segments of proteins are covered by a major N protein which plays a significant role in the virus regulation cycle, including translation, trafficking, as well as assembly. [18]
Although, current evidence from literature-based studies has emphasized that the N protein upon interaction with host cells slowly modulates the host immune response towards an infection. [19] The maturation of Bunyavirus/Orthohantavirus glycoproteins takes place in the Golgi complex. During the process of maturation, Gn and Gc glycoproteins are N-glycosylated. Gn glycoprotein plays a crucial role in supporting Golgi trafficking. The Gn glycoprotein expression for instance it occurs solely, results in partial localization of Golgi. Moreover, when Gc expression occurred then the localization occurs in the endoplasmic reticulum. [20]
Role of Glycoprotein-Induced Virulence Mechanism
Nevertheless, any human body during the entry of viral particles generates the innate immune response which is triggered towards the reduction of viral replication via the (IFNs) Type I interferons which play a significant role in affording direct antiviral protection by activating natural killer (NK) cells. Furthermore, for surveillance inside the host cellular membrane viruses develop regulatory mechanisms that prevent elimination by inhibiting pathway activating type I IFN transcription. [21] It has been demonstrated in several studies that expression of the G1 protein cytoplasmic tail of pathogenic hantaviruses tries to inhibit the induction of IFN-β by binding to TRAF3 and thereby preventing RIG-I/TBK1- directed IRF3 phosphorylation. It has been distinctively displayed in the schematic diagram as shown in (Figure-4) that TRAF3 is an E3 ubiquitin ligase which responsible for the formation of a TBK1– TRAF3 complex, crucial for IRF3 phosphorylation. Hence, viruses are smarter and the N-glycoproteins mimics normal non-pathogenic protein and subsequently interfere with antiviral activity in infected cells, thereby promoting viral replication.
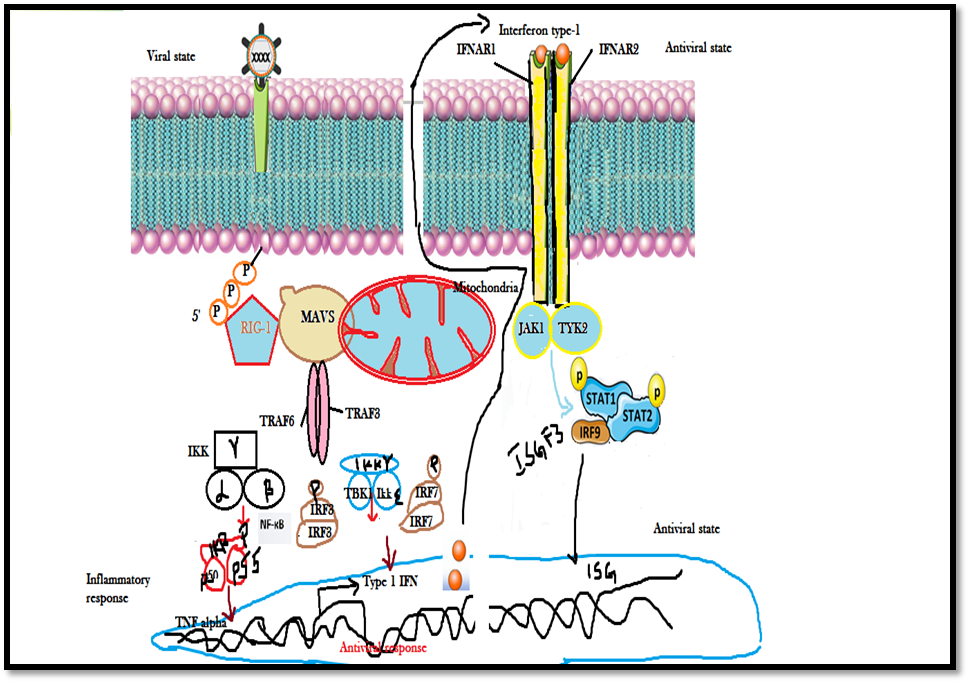
Figure 4: Schematic diagram illustrating the transcriptional activation of Interferon stimulated genes (ISGs) by JAK/STAT signaling pathway
Classification and geographical distribution of Old and New Hantavirus
Earlier, the two important outbursts of disease that happened in the 1950s have eventually led to the detection of Hantaviruses in the Old and New Worlds. The first outbursts happened during the Korean War in the 1950s wherein greater than 3,000 UN troops had fallen sick with Korean hemorrhagic fever, regarded commonly as hemorrhagic fever with renal syndrome (HFRS).[22] The next outbreak of the disease in 1993 seems to occur in the US at four major corners, which was identified with a clinical syndrome called Hantavirus pulmonary syndrome (HPS) or Hantavirus cardiopulmonary syndrome (HCPS). These outbursts of viruses increased the mortality rates in humans approximately 12% (HFRS) and majorly 60% (HPS) in associated diseases. The disease-causing infectious agent was found to be the Hantaan virus (HTNV) transmitted from the striped field mouse (Apodemus agrarius) which was the major reservoir.[23] Later, several other HFRS-related viruses happened in several regions of Asia, Europe, and the United States.
Investigation reports revealed the presence of HTNV and HTNV-like viruses in urban cases leading to HFRS caused by Apodemus agrarius as well as A. peninsulae and Seoul virus (SEOV) rodents which were distributed in Far East Russia, China, Asia, and South Korea in the early 1980s as shown in (Table-1). Other critical reports on a different virus such as Dobrava virus (DOBV), and Dobrava-like viruses caused by old-world Hantaan virus belonging to the genera Apodemus which outbursts in Europe. Nevertheless in Europe, nephropathia epidemica (NE), which is a milder form of HFRS portrayed in the 1930s [24], was found to be caused by another hantavirus, Puumala virus (PUUV), present in bank vole, Myodes glareolus earlier recognized as Clethrionomys glareolus.
The discovery of these Hantaviruses outbreaks causing HFRS has led awareness among the worldwide, especially increasing of this disease in China approximately to 150,000 each year. In contrast to these, Sin Nombre virus (SNV), New world virus belonging to the subfamily Sigmodontinae was identified within weeks in regions of the US. [25] Nevertheless, with many technological advancements in recombinant technology robust detection of this newly discovered virus was possible.
Currently, over 29 hantaviruses have been recognized which cause hemorrhage illness These ailments are transmitted from their rodent reservoirs to humans across worldwide wherever rodent hosts exist (Table 1). In the Middle East, Africa as well as Asia Hantaviruses remain existing but yet, undetected reported. Recent evidence with the detection of shrew-borne Hantaviruses around the globe. Until a sound knowledge from seminal discoveries, a new virus identified as the Thottapalayam virus (TPMV), a long-unclassified virus isolated from the Asian house shrew (Suncus murinus). It was recognized as the only acknowledged shrewborne Hantavirus.[26] Distinctively these Hantaviruses have unraveled the attention of research scientists and public health officials for treatments and further to promote public awareness.
Table 1: Geographic distribution of and disease associated with Old World and New World strains of Hantavirus
|
Group& Subfamily |
Viral Strain |
Abbreviation |
Geographic Distribution |
Rodent host |
Associated disease |
|
Old World Murinae |
Hantaan Virus |
HTNV |
China,South Korea,Russia |
Apodemus agrarius |
HFRS |
|
|
Dobrava-Belgrade virus |
DOBV |
Balkans |
Apodemus flavicollis |
HFRS |
|
|
Seoul virus |
SEOV |
Worldwide |
Rattus |
HFRS |
|
Saaremaa virus |
SAAV |
Europe |
Apodemus agrarius |
HFRS |
|
|
Amur virus |
AMRV |
Far East Russia |
Apodemus peninsulae |
HFRS |
|
|
Soochong virus |
SOOV |
South Korea |
Apodemus peninsulae |
HFRS |
|
|
Arvicolinae |
Puumala virus |
PUUV |
Europe, Asia, and the Americas |
Clethrionomys glareolus |
HFRS/NE |
|
|
Khabarovsk virus |
KHAV |
Far East Russia |
Microtus fortis |
HFRS |
|
|
Muju virus |
MUJV |
South Korea |
Myodes regulus |
HFRS |
|
|
Prospect Hill virus |
PHV |
Maryland |
Microtus pennsylvanicus |
HFRS |
|
|
Tula viru |
TULV |
Russia/Europe |
Microtus arvalis |
HFRS |
|
|
Isla Vista virus |
ISLAV |
North America |
Microtus californicus |
HFRS |
|
|
Topografov virus |
TOPV |
Siberia |
Lemmus sibericus |
HFRS |
|
New World Sigmodontinae Or Neotominae |
Sin Nombre virus |
SNV |
North America |
Peromyscus maniculatus |
HPS |
|
|
Monongahela virus |
MGLV |
North America |
Peromyscus leucopus |
HPS |
|
|
New York virus |
NYV |
North America |
Peromyscus leucopus |
HPS |
|
|
Black Creek Canal virus |
BCCV |
North America |
Sigmodon hispidus |
HPS |
|
|
Bayou virus |
BAYV |
North America |
Oryzomys palustris |
HPS |
|
|
Limestone Canyon virus |
LSCV |
North America |
Peromyscus boylii |
HPS |
|
|
Playa de Oro virus |
OROV |
Mexico |
Oryzomys couesi |
HPS |
|
|
Catacamas virus |
CATV |
Honduras |
Oryzomys couesi |
HPS |
|
|
Choclo virus |
CHOV |
Panama |
Oligoryzomys fulvescens |
HPS |
|
|
Calabazo virus |
CALV |
Panama |
Zygodontomys brevicauda |
HPS |
|
|
Rio Segundo virus |
RIOSV |
Cost Rica |
Reithrodontomys mexicanus |
HPS |
|
|
Cano Delgadito virus |
CADV |
Venezuela |
Sigmodon alstoni |
HPS |
|
|
Andes virus |
ANDV |
Argentina, Chile |
Oligoryzomys longicaudatus |
HPS |
|
|
Bermejo virus |
BMJV |
Argentina |
Oligoryzomys chocoensis |
HPS |
|
|
Pergamino virus |
PRGV |
Argentina |
Akodon azarae |
HPS |
|
|
Lechiguanas virus |
LECV |
Argentina |
Oligoryzomys flavescens |
HPS |
|
|
Maciel virus |
MCLV |
Argentina |
Bolomys obscurus |
HPS |
|
|
Oran virus |
ORNV |
Argentina |
Oligoryzomys longicaudatus |
HPS |
|
|
Alto Paraguay virus |
- |
Paraguayan Chaco |
Holochilus chacoensis |
Zika virus disease in pregnant women |
|
|
Ape Aime virus |
AAIV |
Eastern Paraguay |
Akodon montensis |
Unknown |
|
|
Itapúa virus |
- |
Eastern Paraguay |
Oligoryzomys nigripes |
HPS |
|
|
Rio Mamore virus |
RIOMV |
Bolivia, Peru |
Oligoryzomys microtis |
HPS |
|
|
Araraquara virus |
- |
Brazil |
Bolomys lasiurus |
HPS |
|
|
Juquitiba virus |
JUQV |
Brazil |
Oligoryzomys nigripes |
HPS |
|
|
Jabora´ virus |
JABV |
Brazil, Paraguay |
|
HPS |
Hantavirus Evolution and Analysis
The Hantavirus phylogenetic analyses with their subsequent rodent hosts revealed that they have a long-standing co-evolutionary relationship history with their rodent carriers. The phylogenetic analysis which significantly emphasizes the relationship between the Old World and New world Hanta Virus as shown in (Figure-5) has emphasized the evolution as well as ecological factors that can lead to their emergence and expansion of zoonotic diseases further spillover into human populations. Due to humans overexploitation i.e deforestation, agricultural development, colonization, as well as urbanization have placed human and domestic animal populations at greater threat for various vector-borne diseases. Hence these environmental and ecological changes commonly might reduce rodent diversity but indirectly enhance more host species interaction as well as more hantaviral transmission events within species. Furthermore, this outcome stimulates a cascade of higher transmission of the virus among rodents, leading to a greater threat of viruses among humans.[27]

Figure 5: Phylogenetic analysis for studying the evolutionary relationship between Old and New world Hanta Virus using CLUSTAL (Omega).
Treatment of Hantavirus
Hantaviruses once infect the host cells it robustly triggers an immune response in humans. Nevertheless, the common pathological features of both the old world and New world Hantaviruses results in increased viral infection. Earlier Ribavirin drug has been demonstrated for in vitro and in vivo antiviral activity against members of the Bunyaviridae and the Arenaviridae. This drug reduced the rate of mortality proven to be effective for the treatment of lethal encephalitic suckling mice infected with HTNV. Although several studies were performed in China with HFRS patients revealed the fact that the drug ribavirin through improved prognosis at the early stage of the disease. [28] Yet, when ribavirin is given at the late stage of the disease the patient became anemic had no apparent clinical benefit in curation.
At present there, are no approved post-exposure therapeutic countermeasures against hantaviral infection, but various treatment strategies, which especially target the replication of viral life cycle by increasing the host immunological factors to manage HFRS or HCPS. Several Virus-targeting antivirals drugs, Subunits, antibodies, or even novel small molecules have been employed against Hantavirus infection. Although several antivirals have been proven to be protective in vitro or in vivo, yet, there exist some problems for their clinical application. Several host-targeting medicines have been designed to improve vascular function or rebuild immune homeostasis, while their curative effects are still under debate. Various literature reports on plant-mediated products have been used against HCV but yet no work has been carried on conventional methods for HFRS/HPS recovery.
CONCLUDING REMARKS
This study reveals the evolutionary history of the Hantavirus genus and transmittance due to small mammals such as rodents acting as reservoirs of the virus. However, this review gives a distinct vision on an understanding of Hantavirus evolution and the phylogenetic relationship between the Old World and New world Hantaviruses. Therefore, the emerging threat to the society caused by the upcoming Hantaviruses especially from zoonotic needs to treated and further discovery of novel plant-mediated products with little or no toxicity is a much preferable remedy. Further, future research must also focus on the reduction of this virus from the transmission of rodent hosts to humans.
Contributions
KRP and KRD contributed to writing, drawing figures, and tables in this review article.KRP solely drafted this review article.
Compliance with Ethics Requirements
NIL
ACKNOWLEDGMENT
KRP is thankful to the Department of Biotechnology, Sri Padmavati Mahila Visva Vidyalayam (Women’s) University, Tirupati-India.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
REFERENCES
Mohamed M S, Zohny Y M, El-Senousy W M, Abou A M. Synthesis and Biological Screening of Novel Pyrazoles and their Precursors as Potential Antiviral Agents. Pharmacophores. 2018; 9(1): 126-39.
Qanbarnezhad A, Roustazadeh A, Alizadeh A, Abbastabar H, Nazarnezhad M, Mohseni S. Spatial Distribution of TB and HIV Co-infection in South of Iran. J. Adv. Pharm. Edu. Res. 2018; 8(S2): 51-54.
Alsamarrai A S, Abdulla N H, Aldoori M K. Synthesis and Characterization of 2-((4R, 4aR, 5aS, 6S)-1, 3-dioxo-3, 3a, 4, 4a, 5, 5a, 6, 6a-octahydro-4, 6-ethenocyclopropa [f] isoindol-2 (1H)-yl)-N-(Substituted Phenyl) Acetamides Derivatives Anticipated to Inhibit HIV-1 Activity. Int. J. Pharm. Phytopharm. Res. 2018; 8(5): 7-11.
Rodent-borne diseases. European Centre for Disease Prevention and Control. Retrieved 2018-06-0
Drebot, Jones S.; Grolla, A.; Safronetz, D.; Strong, J. E.; Kobinger, G.; Lindsay, R. L. (4 June 2015). Hantavirus pulmonary syndrome in Canada: An overview of clinical features, diagnostics, epidemiology, and prevention. Canada Communicable Disease Report (Report). Vector-borne diseases in Canada. 41-06. Winnipeg, MB: National Microbiology Laboratory, Public Health Agency of Canada. p. 40. ISSN 1481-8531.
"ICTV 9th Report (2011)–NegativeSenseRNA Viruses – Bunyaviridae". International Committee on Taxonomy of Viruses (ICTV). Retrieved 31 January 2019. Hanta: from Hantaan, river in South Korea near where type virus was isolated.
Spiropoulou, C. F., Goldsmith, C. S., Shoemaker, T. R., Peters, C. J. & Compans, R. W. (2003). Sin Nombre virus glycoprotein trafficking. Virology 308, 48–63.
Jonsson, C. B. & Schmaljohn, C. S. (2001). Replication of hantaviruses. Curr Top Microbiol Immunol 256, 15–32.
Mir, M. A., and Panganiban, A. T. (2004). Trimeric Hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 78, 8281–8288. doi: 10.1128/JVI.78.15.8281-8288.2004.
Matthys, V. S., Cimica, V., Dalrymple, N. A., Glennon, N. B., Bianco, C., and Mackow, E. R. (2014). Hantavirus GnT elements mediate TRAF3 binding and inhibit RIG-I/TBK1-directed beta interferon transcription by blocking IRF3 phosphorylation. J. Virol. 88, 2246–2259. doi: 1128/JVI. 02647-13.
Gil, J., and Esteban, M. (2000). Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): mechanism of action. Apoptosis 5, 107–114. doi: 10.1023/A:1009664109241.
Elliott RM (1990). "Molecular biology of the Bunyaviridae". The Journal of General Virology. 71 (3): 501–522. doi:10.1099/0022-1317-71-3-501.
Mir MA, Panganiban AT (2005). "The Hantavirus Nucleocapsid Protein Recognizes Specific Features of the Viral RNA Panhandle and is Altered in Conformation upon RNA Binding". Journal of Virology. 79 (3): 1824–1835. doi:10.1128/JVI.79.3.1824-1835.2005.
Kielian, M.; Rey, F.A. Virus membrane-fusion proteins: More than one way to make a hairpin. Nat. Rev. Microbiol. 2006, 4, 67–76. 37.
Gavrilovskaya, I.N.; Shepley, M.; Shaw, R.; Ginsberg, M.H.; Mackow, E.R. beta3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA 1998, 95, 7074–7079.
Kim, T.Y.; Choi, Y.; Cheong, H.S.; Choe, J. Identification of a cell surface 30 kDa protein as a candidate receptor for Hantaan virus. J. Gen. Virol. (2002), 83, 767–773.
Mackow, E. R., and I. N. Gavrilovskaya. (2001). Cellular receptors and hantavirus pathogenesis. Curr. Top. Microbiol. Immunol. 256:91–115. 263.
Severson, W., L. Partin, C. S. Schmaljohn, and C. B. Jonsson. (1999). Characterization of the Hantaan nucleocapsid protein-ribonucleic acid interaction. J. Biol. Chem. 274:33732–33739.
Finlay, B. B., and McFadden, G. (2006). Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782. doi: 10.1016/j.cell.2006.01.034.
Ruusala, A., Persson, R., Schmaljohn, C. S., and Pettersson, R. F. (1992). Coexpression of the membrane glycoproteins G1 and G2 of Hantaan virus is required for targeting to the Golgi complex. Virology 186, 53–64. doi: 10.1016/0042-6822(92)90060-3.
Alff, P. J., Gavrilovskaya, I. N., Gorbunova, E., Endriss, K., Chong, Y., Geimonen, E., et al. (2006). The pathogenic NY-1 Hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J. Virol. 80, 9676–9686. doi: 10.1128/JVI.00508-506.
Lee, H. W., P.-W. Lee, and K. M. Johnson. (1978). Isolation of the etiologic agent of Korean hemorrhagic fever. J. Infect. Dis. 137:298–308.
Lee, H. W., P. W. Lee, L. J. Baek, C. K. Song, and I. W. Seong. (1981). Intraspecific transmission of Hantaan virus, etiologic agent of Korean hemorrhagic fever, in the rodent Apodemus agrarius. Am. J. Trop. Med. Hyg. 30:1106–1112.
Mustonen, J., J. Partanen, M. Kanerva, K. Pietila, O. Vapalahti, A. Pasternack, and A. Vaheri. (1996). Genetic susceptibility to severe course of nephropathia epidemica caused by Puumala hantavirus. Kidney Int. 49: 217–221.
Hjelle, B., S. Jenison, N. Torrez-Martinez, B. Herring, S. Quan, A. Polito, S. Pichuantes, T. Yamada, C. Morris, F. Elgh, H. W. Lee, H. Artsob, and R. Dinello. (1997). Rapid and specific detection of Sin Nombre virus antibodies in patients with hantavirus pulmonary syndrome by a strip immunoblot assay suitable for field diagnosis. J. Clin. Microbiol. 35:600–608.
Song, J. W., L. J. Baek, C. S. Schmaljohn, and R. Yanagihara. (2007). Thottapalayam virus, a prototype shrewborne hantavirus. Emerg. Infect. Dis. 13:980–985.
Mills, J. (2006). Biodiversity loss and emerging infectious disease: an example from the rodent-borne hemorrhagic fevers. Biodiversity 7:9–17.
Huggins, J. W., C. M. Hsiang, T. M. Cosgriff, M. Y. Guang, J. I. Smith, Z. O. Wu, J. W. LeDuc, Z. M. Zheng, J. M. Meegan, Q. N. Wang, et al. (1991). Prospective, double-blind, concurrent, placebo-controlled clinical trial of intravenous ribavirin therapy of hemorrhagic fever with renal syndrome. J. Infect. Dis. 164:1119–1127.
