
Applied Science Letters


Mitochondrial DNA CO1 gene sequence was used for the identification of Apis florae(A. florae) which is one of honey bees species, it is called dwarf honey bee as it is small size comparing to other types of honey bees, It makes a single comb on the tree, and was distributed in Asian countries, so it called Asian bees. The dwarf honey bee was recorded for the first time in Africa in 1987 and since that it was recorded in several countries, the dwarf bees are important pollinators. Accidentally A. florae introduced into Saudi Arabia, Jordan, Sudan, Ethiopia, and recently recorded in Egypt (Suez City). A. florae samples were screened for mitochondrial cytochrome oxidase I gene with PCR and the product was sequenced; Sequence of Egyptian Apis florae was analyzed and studied to determine the origin of these population. It was concluded that the Egyptian population was similar and identical with the Indian population by 99% - 100%, query 91-100%, these means that the Egyptian A. florae belong to the Indian species, came from the east, probably from Jordan and move into the Sinai Peninsula, then to Suez city. A sequence of Egyptian population A. florae which is the first record in Egypt was added to the gene bank with Accession No: (PRJNA556448).
The Molecular Origin of the Dwarf Honey Bee Newly Recorded from Egypt
Manal. M. Salem1*, Mohamed A. Shebl1, Soliman M. Kamel1, Hassan A. M. El Demerdash2
1 Depatment of Plant Protection, Faculty of Agriculture, Suez Canal University, Ismailia 41522, Egypt.
2 Department of Food Sciences Technology, Faculty of Environmental Agricultural Sciences, Alaric University, Alarish, Egypt.
ABSTRACT
Mitochondrial DNA COI gene sequence was used for the identification of Apis florae(A. florae) which is one of honey bees species, it is called dwarf honey bee as it is small size comparing to other types of honey bees, It makes a single comb on the tree, and was distributed in Asian countries, so it called Asian bees. The dwarf honey bee was recorded for the first time in Africa in 1987 and since that it was recorded in several coun-tries, the dwarf bees are important pollinators. Accidentally A. florae introduced into Saudi Arabia, Jordan, Sudan, Ethiopia, and recently recorded in Egypt (Suez City). A. florae samples were screened for mitochon-drial cytochrome oxidase I gene with PCR and the product was sequenced; Sequence of Egyptian Apis florae was analyzed and studied to determine the origin of these population. It was concluded that the Egyptian population was similar and identical with the Indian population by 99% - 100%, query 91-100%, these means that the Egyptian A. florae belong to the Indian species, came from the east, probably from Jordan and move into the Sinai Peninsula, then to Suez city. A sequence of Egyptian population A. florae which is the first record in Egypt was added to the gene bank with Accession No: (PRJNA556448).
Keywords: Apis florae, Cytochrome oxidase1 (COI), population genetic, Egypt.
INTRODUCTION
Apis floraeFabricius is widely distributed in Asian countries, southeastern Asia [1, 2]. The honey bee A. florais a wild honey bee belonged to, Family Apidae which is divided into three subfamilies. Apinae, Xylocopinae and Nomadinae. It produces honey, which is a useful food substance [3]. A. florae from tribe Apini [4] It was naturally transported from its local habitat in the east to the west [5]. It is widely distributed in Jordan [6] and Arabian Pensisuila [7], some parts of Africa [5, 8, 9]. The dwarf bee is colonizing many parts of Egypt since it was recorded for the first time in 2017 [10, 11]. With this widespread of the species still, there is no enough data about the molecular originality for each population recorded in each country or region. The Sudanese population was originally introduced from Pakistan [5] which we assume is not the same origin of the Egyptian population. The species has not been recorded so far from the Southern Plateau of Egypt. On the contrary, In Jordan, the populations seem to have 2 origins linked to one population from Sudan, Pakistan, Oman, Iran, and Saudi Arabia, The second one is related to Sri Lankans populations and southern Indian [6]. It is suggested that Sudanese and Jordanian populations could move to Sinai Peninsula [7, 12], but some expeditions have been conducted without any single record of the species.
There are no subspecies of A. florae that have been reported worldwide due to the inability of A. florae to disperse from peripheral populations which lead to limited regional differentiation of these species into recognizable subspecies/races [7]. Here in this paper, the Egyptian population was subjected to studying their molecular originality using Mitochondrial DNA Cytochrome oxidase1 (COI). The current study aims to identify and detect the origin of newly recorded A. florae in Egypt, collected from the Suez Canal University campus nest received from Suez Gulf in (2017) and Suez City in October (2017 to 2018) during the activity of bees in the morning from Basil flowers. Mitochondrial DNA Cytochrome oxidase1 (COI) was used to identify species. Cytochrome oxidase1 gene is a well-defined gene which is unique enough to identify species [13, 14].
To achieve this work several trips were carried out to collect A. florae samples from Suez City from October 2017 to October 2018 and the samples were identified to isolate 690 pp. DNA of the COI gene and the gene were analyzed and phylogenetic trees were constructed.
MATERIAL AND METHODS
Specimen's collection
Specimens were collected using sweep net and preserved life in plastic jars. The Egyptian population was collected from Suez through several fieldworks conducted in the city from 2017 to 2018.
DNA-extraction, Amplification, and Sequencing
DNA has been extracted from legs after grinding tissue well with digestion solution (lysis buffer) using the protocol for animal tissues of DNeasy tissue kit (QIAGEN) we extracted The DNA of bee samples in Biotechnology Research Institute lab in Suez Canal University the DNA of bee samples were extracted successfully. Concentration and purity of DNA of each sample was measured by using Nanodrop photometer. Some samples were used for the DNA extraction but 4 samples were subjected for DNA sequencing (3 samples representing the Egyptian population.
The selected primers used in this study were one Forward (Barbee CAACAAATCATAAAAATATTGG, and one revers MTD9 CCCGGTAAAATTAAAATATAAACTTC, in two directions according to (Cytochrome oxidase I gene for Corbiculate bees) [15].
The total DNA obtained from each specimen was diluted to 50 ng /μl and used as template DNA.
PCR reaction usually contains 2 primers, one binds in the forward" Orientation to target DNA sequence 5' end, and the other binds to the "reverse" orientation of the target 3' end. Primers were obtained from Matrix Company and diluted with deionized water to become its concentration 20 pk.in PCR reactions we used 2.0 µl of DNA template in a 25 µl final volume containing 12,5 PCR buffer (MM), 1 µl each primer (20) pm. The condition for PCR was the following: initial denaturation step at 95 °C for 5 min; followed by 35Cycles at 95 °C for 1 min, 50 °C for 1 min; and extension at 72°C for 5 min, and cooling to 4°C.
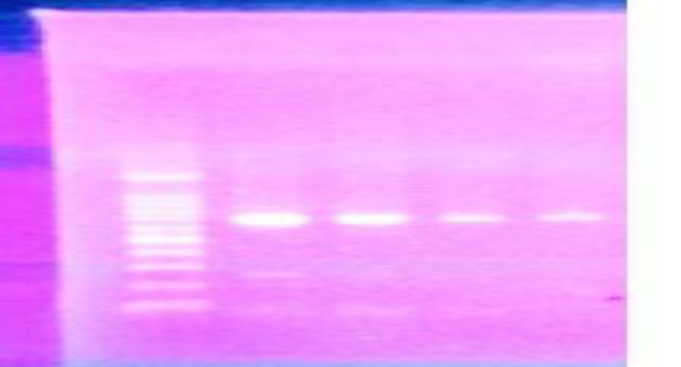
Figure 1: Gel electrophoresis of specific bands for Egyptian samples of A. Florae COI gene at 690 pp.
PCR products were separated on a 1.5% agarose gel using 10 μl PCR product, the ladder used was 100 bp, stained using Gel Red 10.000X (Biotium, USA) and have seen under UV light, it has been purified using 0.5 µl of Exo SA P-IT (USB Corporation) according to the recommendations of the manufacturer and has been sequenced from both directions (Macrogen Company, South Korea). PCR product was used in two directions for sequencing. The genetic variability of the COI barcode region was compared. COI sequences have been edited and translated into amino acid sequences. Finally, the sequences were analyzed using the BLAST tool against Gen Bank [16] and BOLD [17] databases characters morphology of A.florae.
RESULTS
Cytochrome oxidase gene of three samples A. florae samples from the Ismailia governorate has been sequenced. The identity and divergence of samples with others in the gene bank were compared. Sequences of A. florae from Egypt (Suez City) agreed with the sequences of Indian with 99 to 100%, query 93 to100%, the similarity to Pakistanis bee with Identification with 100%, but with a query (81-83) lower than Indian bee and the similarity to Iran bee with lower Identification (99.33% to 99.65%) and lower query (86%-87%) than Indian bee.
Table 1: Identity and divergence of newly recorded Egyptian A. florae with others in the Middle East and the worldwide
|
origin |
s 3 |
s 2 |
Sample 1 |
Accession (N) |
|||
|
Q |
I |
Q |
I |
Query |
Identity |
||
|
India |
92% |
99.84% |
91% |
100% |
92% |
100% |
GenBank KT960841.1 |
|
India |
92% |
99.83% |
92% |
99,54% |
93% |
99,89% |
GenBank KR010698.1 |
|
India. |
92% |
99, 69% |
91% |
99,65% |
91% |
99,85% |
GenBank KU737494. 1 |
|
India. |
92% |
99,53% |
83% |
99,56 |
92% |
99,54 |
GenBankKU666428.l |
|
Bangladish |
91% |
99.22% |
86% |
99.55% |
91% |
99.38% |
GenBank MH378769.1 |
|
Pakistan |
81% |
100% |
91% |
99,69 |
81% |
100% |
GenBank: KY844742.1 |
|
Iran |
87% |
99,33% |
86% |
99,35 |
86% |
99,96% |
GenBank: MG548258.1 |

Figure 2: Similarity of nucleotides of A.florae from Egypt showed that very identical with divergence 100% percent with the nucleotides of India population Accession No (KT960841.1)

Figure 3: Phylogenetic tree for the relationship between A. florae from Egypt with the Middle East, it showed that A. florae from Egypt are similar and identical with Indian species with percentage (100%).
DISCUSSION
The molecular analysis based on DNA barcoding proved that the Egyptian population is much closed and identical with the Indian population, we used mitochondrial cytochrome oxidase I gene.
sequences of A. florae from Egypt (Suez City) agreed with the sequences of Makk (2015) [17] (Pollinators of Punjab, India) Molecular characterization and DNA barcoding of Insects collected from Kass Plateau with 99.54% to 100% [18], query 93%to100 % the similarity to Pakistanis bee with Identification 100%, but with a query (81-83) lower than Indian bee and the similarity to Iran bee with lower Identification (99.65 % to 99.33%) and lower query (86%-87%) than Indian bee (table 1) this means that the Egyptian Population is more similar to the Indian population than Pakistanis then Iran population, especially that the bodyweight of the Egyptian population A. florae bee was (17 to 31 mg) we measured weight eleven samples from Egyptian population the average weight was equal with a weight of Indian population bee in the previous study [19].
Morphological Determination of Egyptian population origin comparing to the neighboring populations in Saudi Arabia and Jordan is an important step [10]. However, body size and most morphometric characteristics of dwarf honey bee (A. florae) varied significantly depending on the geographical origin. In Saudi Arabia, most dwarf bees measured characteristics values were lower than those reported in Nepal, Iran, Pakistan, Myanmar, India, and Thailand, while they were higher than those reported from Sri Lanka [20].
Mitochondrion DNA COI Sequnce of Egyptian population A. florae samples are similar and identical with the second Origen linked to the Indian population. Based on all the previous, we concluded that the newly recorded Egyptian population A. florae belonged to Indian population and came from the east, probably from Jordan over Aqaba Gulf or from India itself during transporting, anyway It is highly unlikely that he will come from Saudi Arabia because the Egyptian population A. florae samples identical with Indian and Pakistanis population with 100%, Indian and Pakistanis population are higher than those reported from Saudi Arabia [20].
CONCLUSION
The newly recorded Egyptian population A. florae belonged to the Indian population and came from the east, probably from Jordan over Aqaba Gulf or from India itself during transporting; we need more studies to detect where did Egyptian population A. florae came from.
ACKNOWLEDGMENTS
The authors are grateful to research facilities offered by the Biotechnology Research Institute, Suez Canal University for performing the DNA extraction. We are grateful to Prof. Dr. Mohamed El Shahidy and, Faculty of Veterinary Medicine, Suez Canal University during for his help and support during the study.
REFERENCES
